A large-scale volumetric correlated light and electron microscopy study localizes Alzheimer's disease-related molecules in the hippocampus
bioRxiv, 2023.
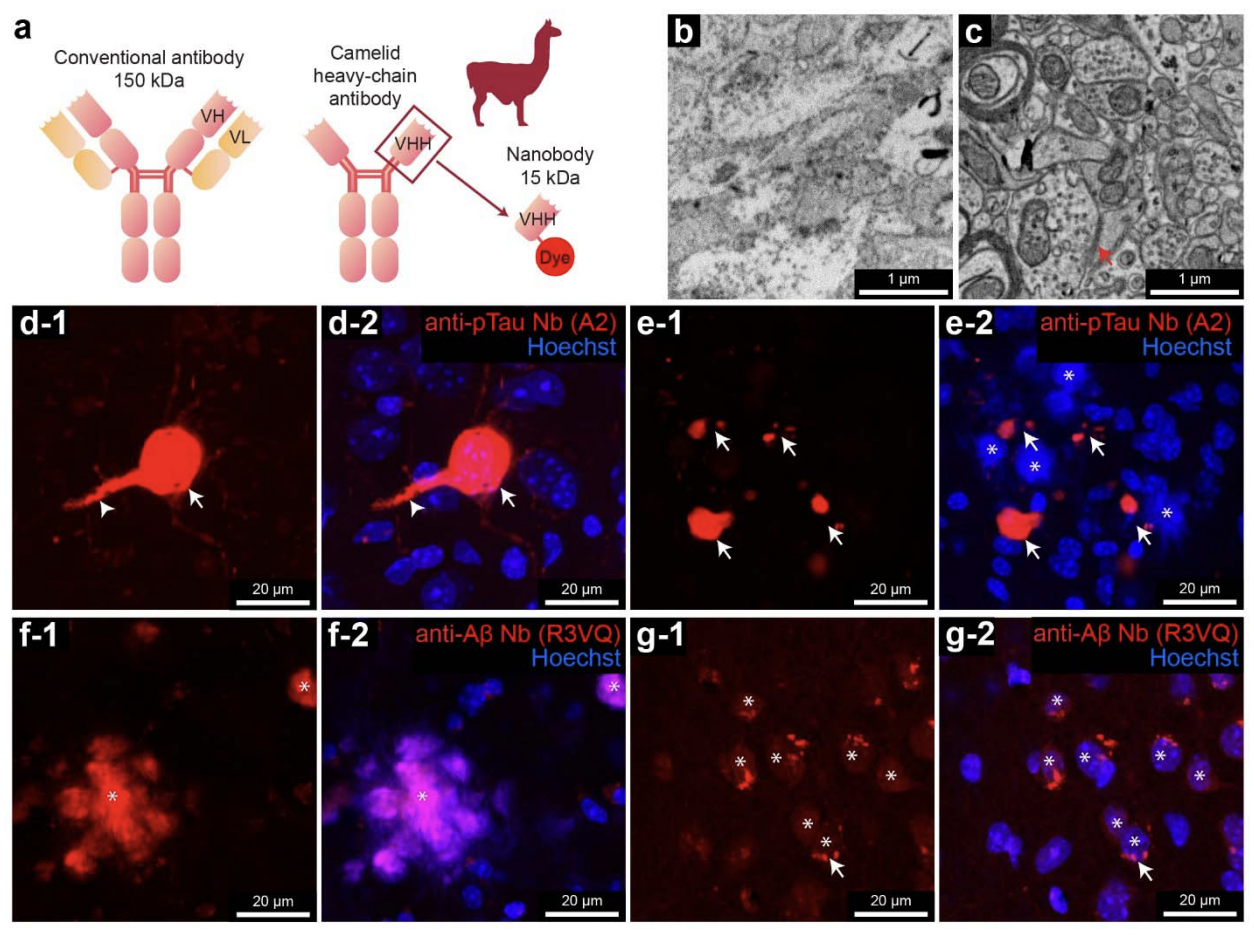
Connectomics is a nascent neuroscience field to map and analyze neuronal networks. It provides a new way to investigate abnormalities in brain tissue, including in models of Alzheimer's disease (AD). This age-related disease is associated with alterations in amyloid- (A ) and phosphorylated tau (pTau). These alterations correlate with AD's clinical manifestations, but causal links remain unclear. Therefore, studying these molecular alterations within the context of the local neuronal and glial milieu may provide insight into disease mechanisms. Volume electron microscopy (vEM) is an ideal tool for performing connectomics studies at the ultrastructural level, but localizing specific biomolecules within large-volume vEM data has been challenging. Here we report a volumetric correlated light and electron microscopy (vCLEM) approach using fluorescent nanobodies as immuno-probes to localize Alzheimer’s disease- related molecules in a large vEM volume. Three molecules (pTau, A , and a marker for activated microglia (CD11b)) were labeled without the need for detergents by three nanobody probes in a sample of the hippocampus of the 3xTg Alzheimer’s disease model mouse. Confocal microscopy followed by vEM imaging of the same sample allowed for registration of the location of the molecules within the volume. This dataset revealed several ultrastructural abnormalities regarding the localizations of A and pTau in novel locations. For example, two pTau-positive post-synaptic spine-like protrusions innervated by axon terminals were found projecting from the axon initial segment of a pyramidal cell. Three pyramidal neurons with intracellular A or pTau were 3D reconstructed. Automatic synapse detection, which is necessary for connectomics analysis, revealed the changes in density and volume of synapse at different distances from an A plaque. This vCLEM approach is useful to uncover molecular alterations within large-scale volume electron microscopy data, opening a new connectomics pathway to study Alzheimer’s disease and other types of dementia.