AxonEM Dataset: 3D Axon Instance Segmentation of Brain Cortical Regions
arXiv preprint arXiv:2107.05451 (MICCAI), 2021.
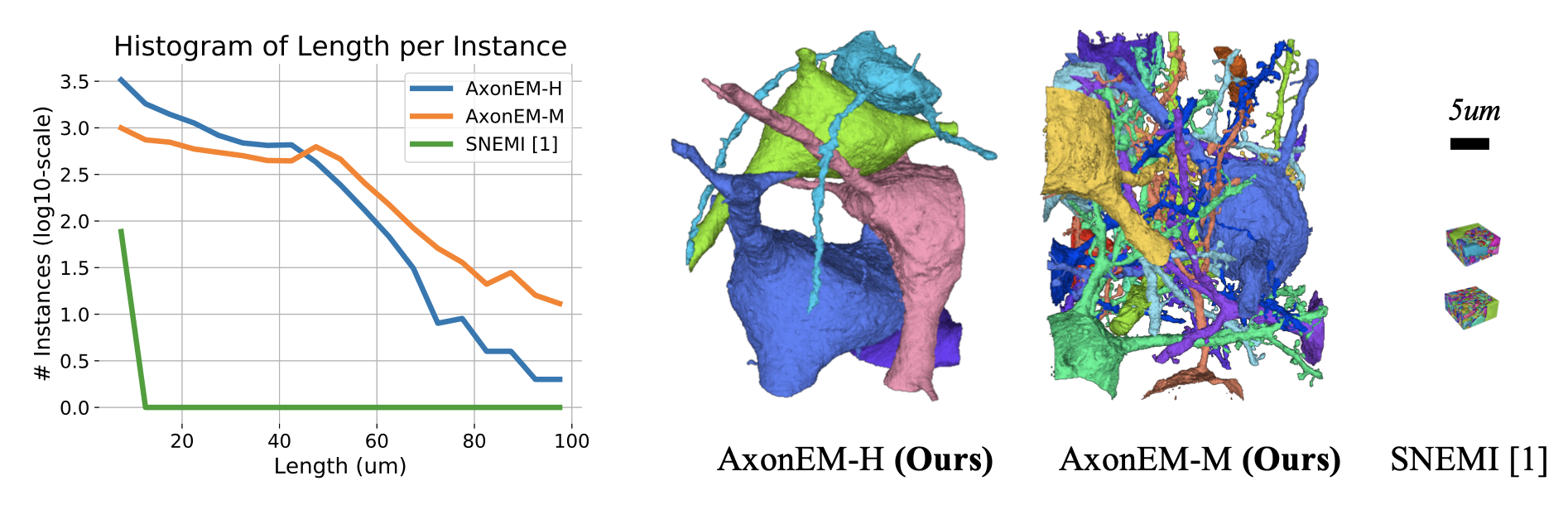
Electron microscopy (EM) enables the reconstruction of neu- ral circuits at the level of individual synapses, which has been transforma- tive for scientific discoveries. However, due to the complex morphology, an accurate reconstruction of cortical axons has become a major chal- lenge. Worse still, there is no publicly available large-scale EM dataset from the cortex that provides dense ground truth segmentation for axons, making it difficult to develop and evaluate large-scale axon reconstruc- tion methods. To address this, we introduce the AxonEM dataset, which consists of two 30×30×30 μm3 EM image volumes from the human and mouse cortex, respectively. We thoroughly proofread over 18,000 axon in- stances to provide dense 3D axon instance segmentation, enabling large- scale evaluation of axon reconstruction methods. In addition, we densely annotate nine ground truth subvolumes for training, per each data vol- ume. With this, we reproduce two published state-of-the-art methods and provide their evaluation results as a baseline. We publicly release our code and data at https://connectomics-bazaar.github.io/proj/ AxonEM/index.html to foster the development of advanced methods.