Current Progress and Challenges in Large-scale 3D Mitochondria Instance Segmentation
IEEE Transactions on Medical Imaging (IEEE TMI), 2023.
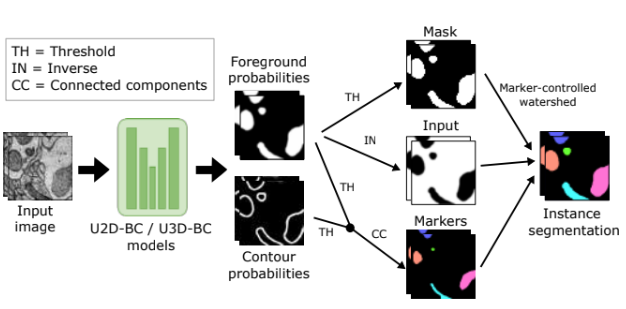
In this paper, we present the results of the MitoEM challenge on mitochondria 3D instance segmentation from electron microscopy images, organized in conjunction with the IEEE-ISBI 2021 conference. Our benchmark dataset consists of two large-scale 3D volumes, one from human and one from rat cortex tissue, which are 1,986 times larger than previously used datasets. At the time of paper submission, 257 participants had registered for the challenge, 14 teams had submitted their results, and six teams participated in the challenge workshop. Here, we present eight top-performing approaches from the challenge participants, along with our own baseline strategies. Posterior to the challenge, annotation errors in the ground truth were corrected without altering the final ranking. Additionally, we present a retrospective evaluation of the scoring system which revealed that (1) the challenge metric was permissive with the false positive predictions and (2) the size-based grouping of instances did not correctly categorize mitochondria of interest. Thus, we propose a new scoring system that better reflects the correctness of the segmentation results. Although several of the top methods are compared favorably to our own baselines, substantial errors remain unsolved for mitochondria with challenging morphologies. Thus, the challenge remains open for submission and automatic evaluation, with all volumes available for download.
Acknowledgements
We gratefully thank the Grand Challenge team for providing the platform that enables public access, challenge organization, and automatic evaluation. This work has been partially supported by NSF award IIS-1835231, NIH award 5U54CA225088-03, by the University of the Basque Country UPV/EHU grant GIU19/027, by Ministerio de Ciencia, Innovacion y Universidades, MCIN/AEI/10.13039/501100011033, ́under grant PID2021-126701OB-I00, by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001999), the UK Medical Research Council (FC001999), and the Wellcome Trust (FC001999), by the Frederick National Laboratory for Cancer Research under NIH Contract No. 75N91019D00024.