Instance Segmentation of Unlabeled Modalities via Cyclic Segmentation GAN
arXiv preprint arXiv:2204.03082, 2022.
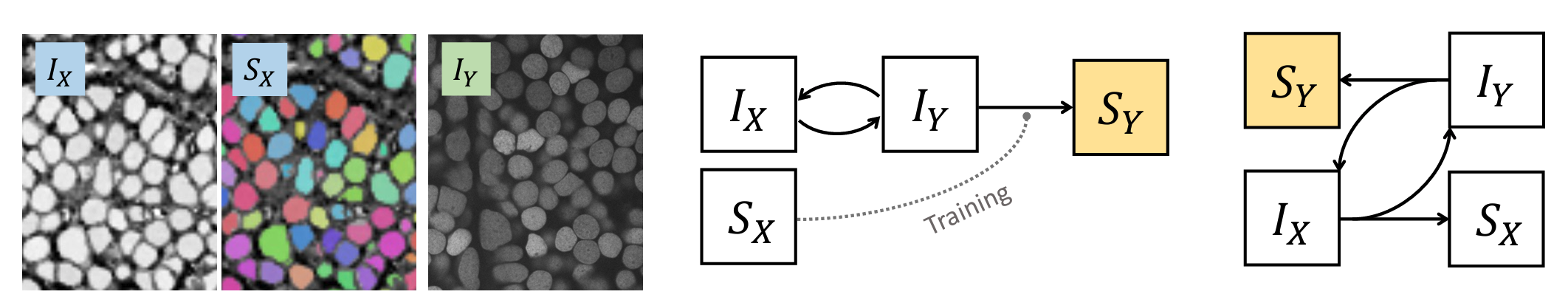
Instance segmentation for unlabeled imaging modalities is a challenging but essential task as collecting expert annotation can be expensive and time-consuming. Existing works segment a new modality by either deploying a pre-trained model optimized on diverse training data or conducting domain translation and image segmentation as two independent steps. In this work, we propose a novel Cyclic Segmentation Generative Adversarial Network (CySGAN) that conducts image translation and instance segmentation jointly using a unified framework. Besides the CycleGAN losses for image translation and supervised losses for the annotated source domain, we introduce additional self-supervised and segmentation-based adversarial objectives to improve the model performance by leveraging unlabeled target domain images. We benchmark our approach on the task of 3D neuronal nuclei segmentation with annotated electron microscopy (EM) images and unlabeled expansion microscopy (ExM) data. Our CySGAN outperforms both pretrained generalist models and the baselines that sequentially conduct image translation and segmentation. Our implementation and the newly collected, densely annotated ExM nuclei dataset, named NucExM, are available.
Acknowledgements
This work has been partially supported by NSF awards IIS-1835231 and IIS2124179 and NIH grant 5U54CA225088-03. Leander Lauenburg acknowledges the support from a fellowship within the IFI program of the German Academic Exchange Service (DAAD). Ignacio Arganda-Carreras acknowledges the support of the Beca Leonardo a Investigadores y Creadores Culturales 2020 de la Fundaci´on BBVA. Edward S. Boyden acknowledges NIH 1R01EB024261, Lisa Yang, John Doerr, NIH 1R01MH123403, NIH 1R01MH123977, Schmidt Futures.