Exploring the Connectome - Petascale Volume Visualization of Microscopy Data Streams
IEEE Computer Graphics and Applications, 2013.
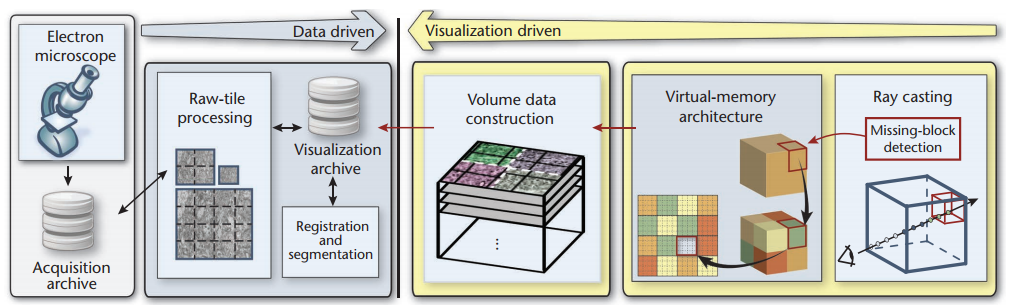
Recent advances in high-resolution microscopy allow neuroscientists to acquire volume data of neural tissue of extreme size. However, the tremendous resolution and the high complexity of neural structures present big challenges to storage, processing, and visualization at interactive rates. We present a system for interactive exploration of petascale (petavoxel) volumes resulting from high-throughput electron microscopy data streams. Our system can concurrently handle multiple volumes, and also supports the simultaneous visualization of high-resolution voxel segmentation data. We employ a visualization-driven system design that allows us to restrict most computations to a small sub-set of the data. We employ a multi-resolution virtual memory architecture for better scalability than previous approaches and handling of incomplete data. We illustrate the real-world use of our system for a mouse cortex volume of one teravoxel in size, where several hundred neurites as well as synapses have been segmented and labeled.