Interactive Volume Exploration of Petascale Microscopy Data Streams Using a Visualization-Driven Virtual Memory Approach
IEEE Transactions on Visualization and Computer Graphics (SciVistextquoteright12), 2012.
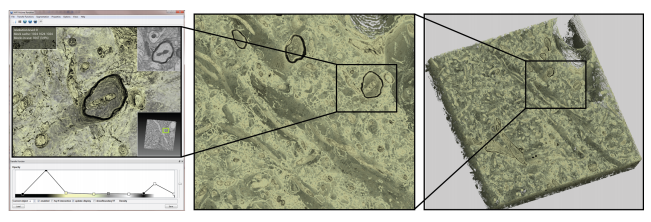
This paper presents the first volume visualization system that scales to petascale volumes imaged as a continuous stream of high-resolution electron microscopy images. Our architecture scales to dense, anisotropic petascale volumes because it: (1) decouples construction of the 3D multi-resolution representation required for visualization from data acquisition, and (2) decouples sample access time during ray-casting from the size of the multi-resolution hierarchy. Our system is designed around a scalable multi-resolution virtual memory architecture that handles missing data naturally, does not pre-compute any 3D multi-resolution representation such as an octree, and can accept a constant stream of 2D image tiles from the microscopes. A novelty of our system design is that it is visualization-driven: we restrict most computations to the visible volume data. Leveraging the virtual memory architecture, missing data are detected during volume ray-casting as cache misses, which are propagated backwards for on-demand out-of-core processing. 3D blocks of volume data are only constructed from 2D microscope image tiles when they have actually been accessed during ray-casting. We extensively evaluate our system design choices with respect to scalability and performance, compare to previous best-of-breed systems, and illustrate the effectiveness of our system for real microscopy data from neuroscience.