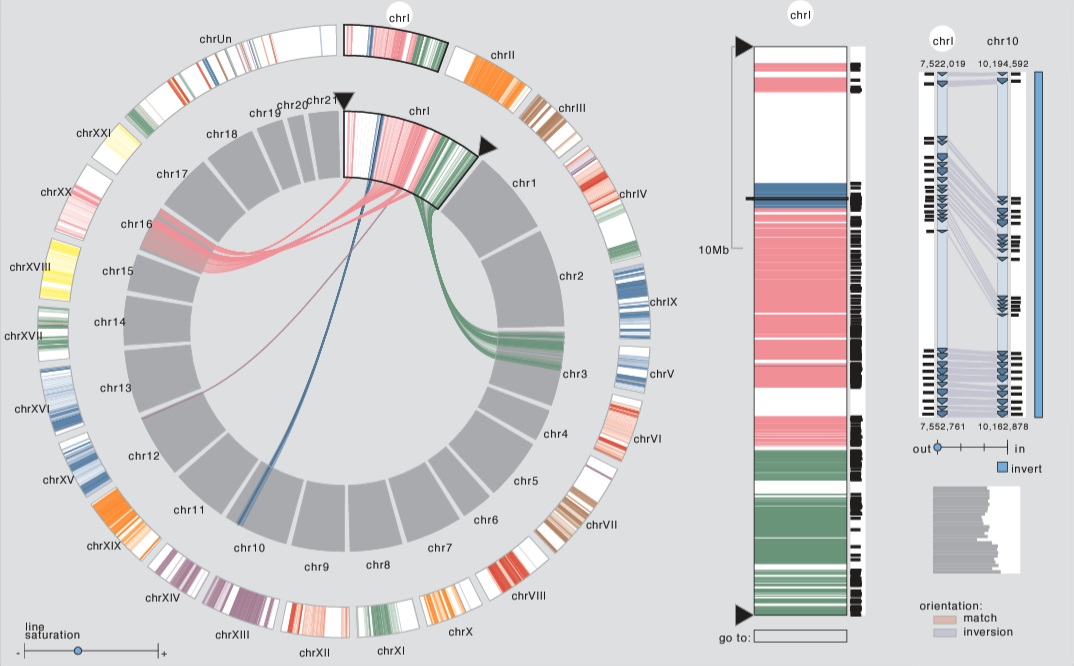
MizBee: A Multiscale Synteny Browser
IEEE transactions on visualization and computer graphics, 2009.
In the field of comparative genomics, scientists seek to answer questions about evolution and genomic function by comparing the genomes of species to find regions of shared sequences. Conserved syntenic blocks are an important biological data abstraction for indicating regions of shared sequences. The goal of this work is to show multiple types of relationships at multiple scales in a way that is visually comprehensible in accordance with known perceptual principles. We present a task analysis for this domain where the fundamental questions asked by biologists can be understood by a characterization of relationships into the four types of proximity/location, size, orientation, and similarity/strength, and the four scales of genome, chromosome, block, and genomic feature. We also propose a new taxonomy of the design space for visually encoding conservation data. We present MizBee, a multiscale synteny browser with the unique property of providing interactive side-by-side views of the data across the range of scales supporting exploration of all of these relationship types. We conclude with case studies from two biologists who used MizBee to augment their previous automatic analysis work flow, providing anecdotal evidence about the efficacy of the system for the visualization of syntenic data, the analysis of conservation relationships, and the communication of scientific insights.
Acknowledgements
This work was funded in part by the Initiative in Innovative Computing at Harvard University. We thank our biology collaborators, Manfred Grabherr and Li-Jun Ma from the Broad Institute, for their time and the use of their data, and Janet Iwasa and Matthew Tobiasz for their helpful comments regarding previous drafts of this paper. We also thank the Broad Viz Group for seeding the initial inspiration of this work, and for continued feedback throughout the development.