NucMM Dataset: 3D Neuronal Nuclei Instance Segmentation at Sub-Cubic Millimeter Scale
, 2021.
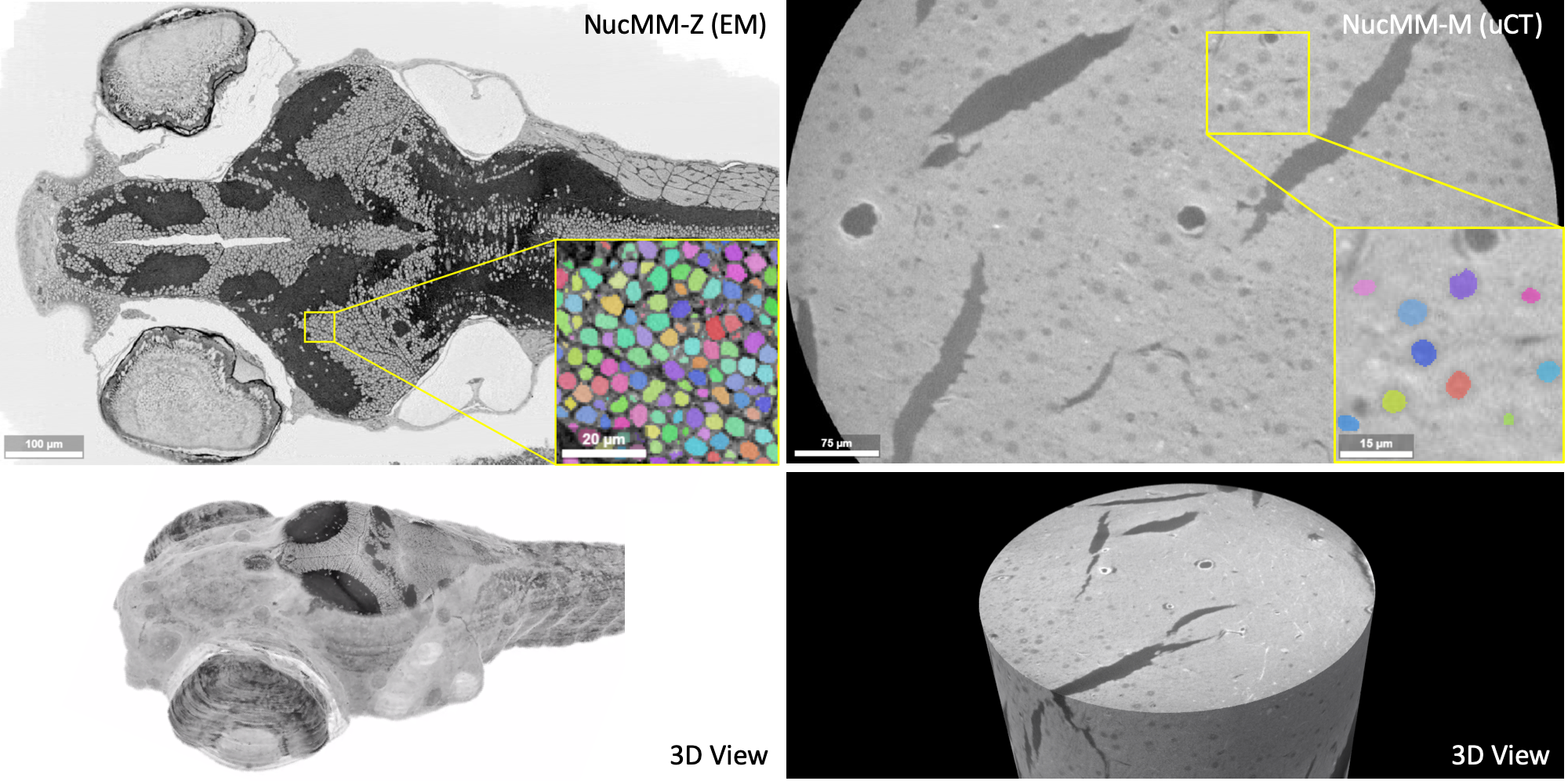
Segmenting 3D cell nuclei from microscopy image volumes is critical for biological and clinical analysis, enabling the study of cellular expression patterns and cell lineages. However, current datasets for {em neuronal} nuclei usually contain volumes smaller than $10^{text{-}3} mm^3$ with fewer than 500 instances per volume, unable to reveal the complexity in large brain regions and restrict the investigation of neuronal structures. In this paper, we have pushed the task forward to the sub-cubic millimeter scale and curated the {em NucMM} dataset with two fully annotated volumes: one $0.1 mm^3$ electron microscopy (EM) volume containing nearly the entire zebrafish brain with around 170,000 nuclei; and one $0.25 mm^3$ micro-CT (uCT) volume containing part of a mouse visual cortex with about 7,000 nuclei. With two imaging modalities and significantly increased volume size and instance numbers, we discover a great diversity of neuronal nuclei in appearance and density, introducing new challenges to the field. We also perform a statistical analysis to illustrate those challenges quantitatively. To tackle the challenges, we propose a novel hybrid-representation learning model that combines the merits of foreground mask, contour map, and signed distance transform to produce high-quality 3D masks. The benchmark comparisons on the NucMM dataset show that our proposed method significantly outperforms state-of-the-art nuclei segmentation approaches. Code and data are available at url{https://connectomics-bazaar.github.io/proj/nucMM/index.html}.