PyTorch Connectomics: A Scalable and Flexible Segmentation Framework for EM Connectomics
, 2021.
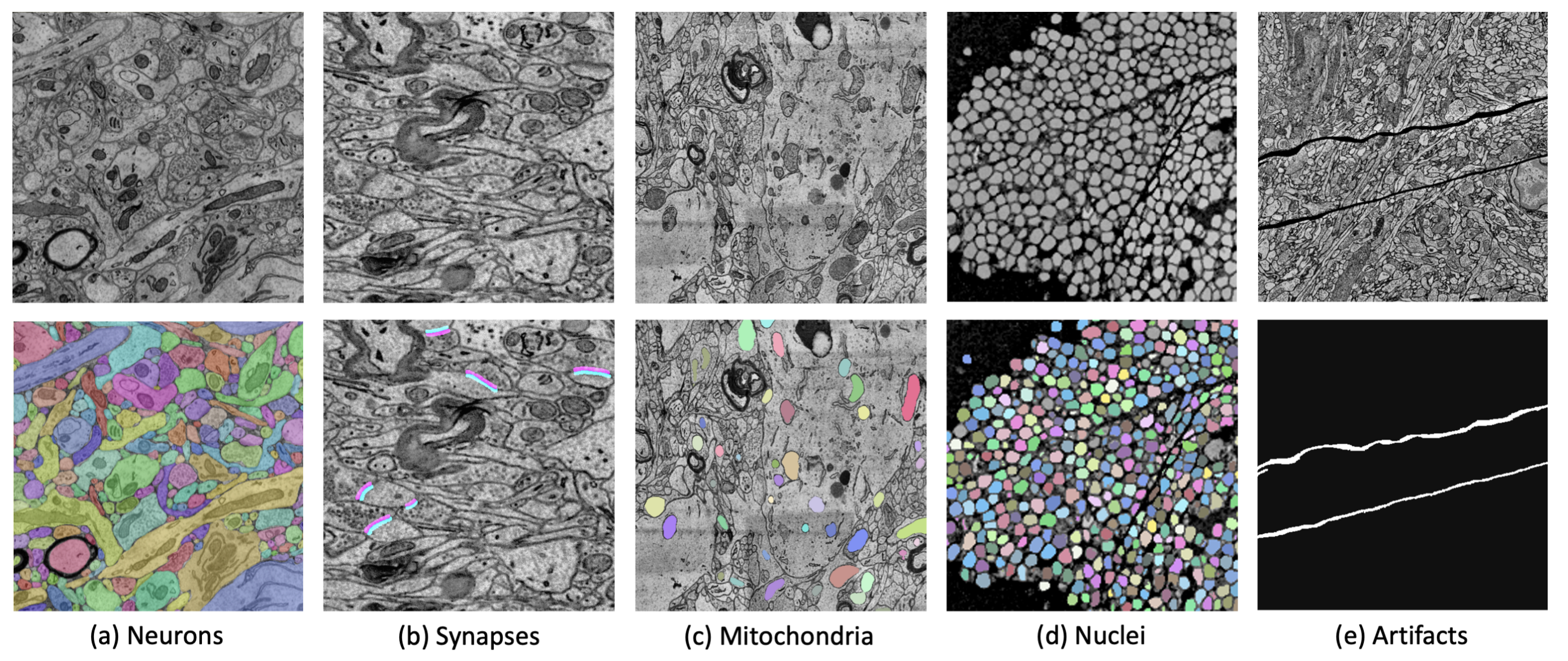
We present PyTorch Connectomics (PyTC), an open-source deep-learning framework for the semantic and instance segmentation of volumetric microscopy images, built upon PyTorch. We demonstrate the effectiveness of PyTC in the field of connectomics, which aims to segment and reconstruct neurons, synapses, and other organelles like mitochondria at nanometer resolution for understanding neuronal communication, metabolism, and development in animal brains. PyTC is a scalable and flexible toolbox that tackles datasets at different scales and supports multi-task and semi-supervised learning to better exploit expensive expert annotations and the vast amount of unlabeled data during training. Those functionalities can be easily realized in PyTC by changing the configuration options without coding and adapted to other 2D and 3D segmentation tasks for different tissues and imaging modalities. Quantitatively, our framework achieves the best performance in the CREMI challenge for synaptic cleft segmentation (outperforms existing best result by relatively 6.1%) and competitive performance on mitochondria and neuronal nuclei segmentation.