The XPRESS Challenge: Xray Projectomic Reconstruction -- Extracting Segmentation with Skeletons
Arxiv, 2023.
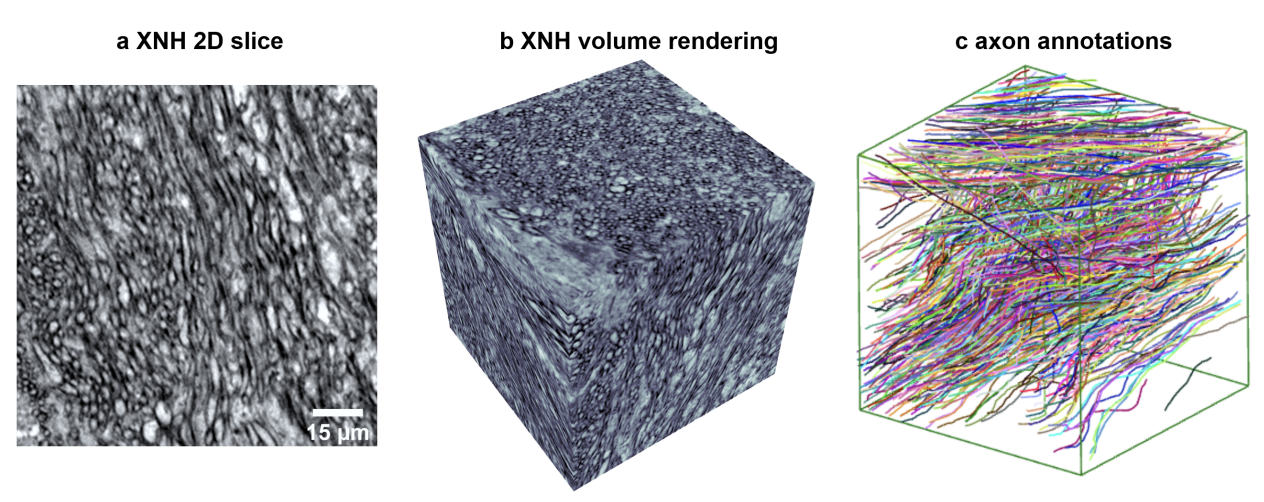
The wiring and connectivity of neurons form a critical structural basis for the function of the nervous system. Advances in volume electron microscopy (EM) and image segmentation have enabled mapping of circuit diagrams (connectomics) within local regions of the mouse brain. However, applying volume EM over the whole brain is not currently feasible due to technological challenges. As a result, comprehensive maps of long-range connections between brain regions are lacking. Recently, we demonstrated that X-ray holographic nanotomography (XNH) can provide high-resolution images of brain tissue at a much larger scale than EM. In particular, XNH is wellsuited to resolve large, myelinated axon tracts (white matter) that make up the bulk of long-range connections (projections) and are critical for inter-region communication. Thus, XNH provides an imaging solution for brain-wide projectomics. However, because XNH data is typically collected at lower resolutions and larger fields-of-view than EM, accurate segmentation of XNH images remains an important challenge that we present here. In this task, we provide volumetric XNH images of cortical white matter axons from the mouse brain along with ground truth annotations for axon trajectories. Manual voxel-wise annotation of ground truth is a time-consuming bottleneck for training segmentation networks. On the other hand, skeleton-based ground truth is much faster to annotate, and sufficient to determine connectivity. Therefore, we encourage participants to develop methods to leverage skeleton-based training. To this end, we provide two types of ground-truth annotations: a small volume of voxel-wise annotations and a larger volume with skeleton-based annotations. The participants will have the flexibility to use either or both of the provided annotations to train their models, and are challenged to submit an accurate voxel-wise prediction on the test volume. Entries will be evaluated on how accurately the submitted segmentations agree with the ground-truth skeleton annotations.