VESICLE: Volumetric Evaluation of Synaptic Interfaces using Computer Vision at Large Scale
BMVA Press: Proceedings of the British Machine Vision Conference (BMVC), 2015.
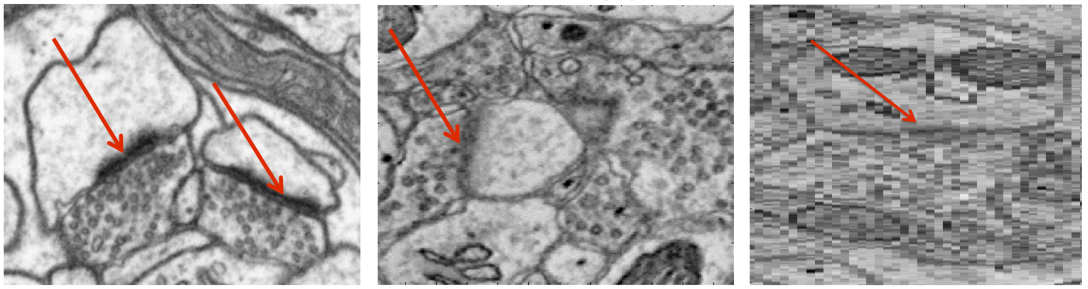
An open challenge at the forefront of modern neuroscience is to obtain a comprehensive mapping of the neural pathways that underlie human brain function; an enhanced understanding of the wiring diagram of the brain promises to lead to new breakthroughs in diagnosing and treating neurological disorders. Inferring brain structure from image data, such as that obtained via electron microscopy (EM), entails solving the problem of identifying biological structures in large data volumes. Synapses, which are a key communication structure in the brain, are particularly difficult to detect due to their small size and limited contrast. Prior work in automated synapse detection has relied upon time-intensive, error-prone biological preparations (isotropic slicing, post-staining) in order to simplify the problem. This paper presents VESICLE, the first known approach designed for mammalian synapse detection in anisotropic, non-poststained data. Our methods explicitly leverage biological context, and the results exceed existing synapse detection methods in terms of accuracy and scalability. We provide two different approaches - a deep learning classifier (VESICLE-CNN) and a lightweight Random Forest approach (VESICLE-RF), to offer alternatives in the performance-scalability space. Addressing this synapse detection challenge enables the analysis of high-throughput imaging that is soon expected to produce petabytes of data, and provides tools for more rapid estimation of brain-graphs. Finally, to facilitate community efforts, we developed tools for large-scale object detection, and demonstrated this framework to find ~50,000 synapses in 60,000 um^3 (220 GB on disk) of electron microscopy data.