A connectomic resource for neural cataloguing and circuit dissection of the larval zebrafish brain
bioRxiv, 2025.
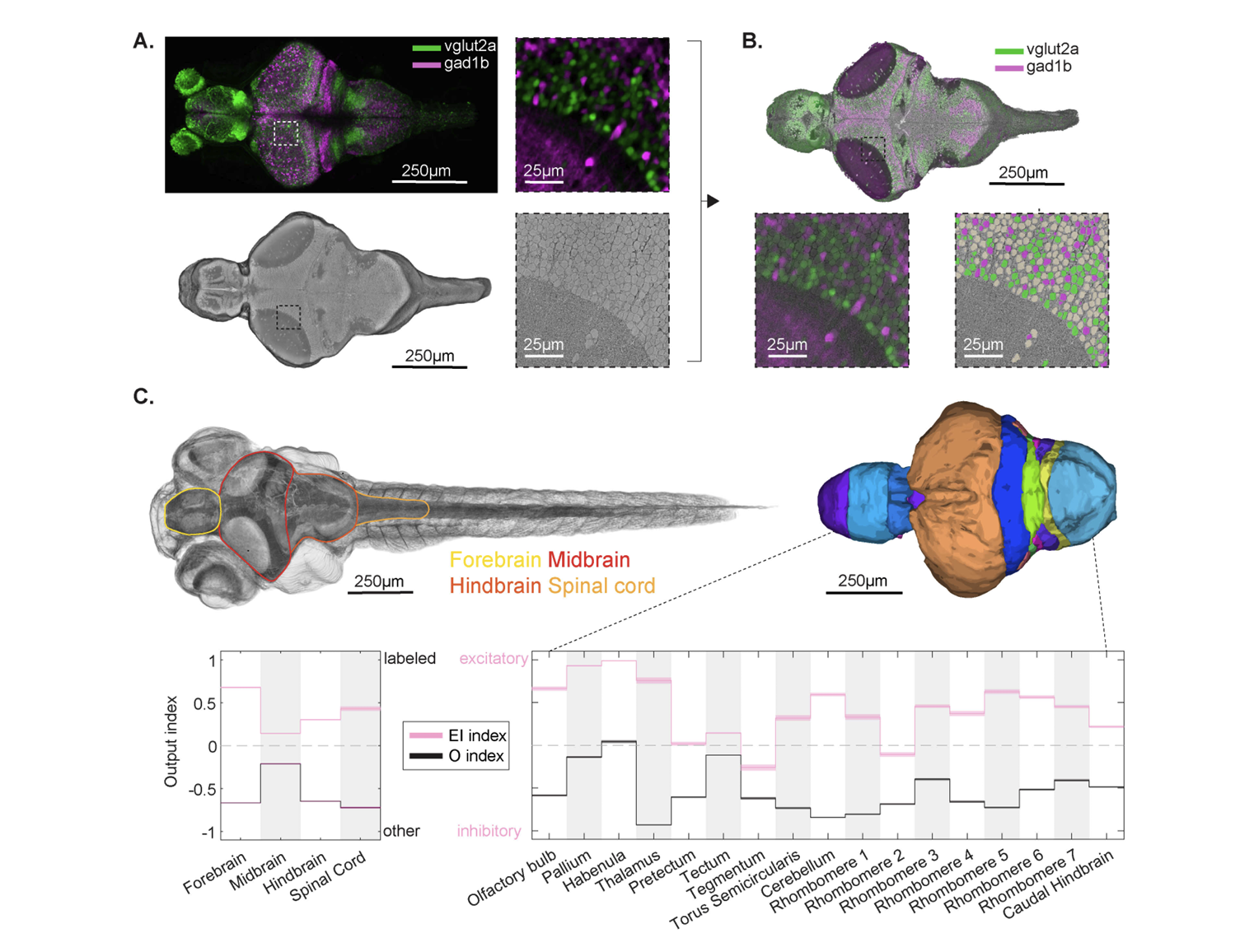
We present a correlated light and electron microscopy (CLEM) dataset from a 7-day-old larval zebrafish, integrating confocal imaging of genetically labeled excitatory (vglut2a) and inhibitory (gad1b) neurons with nanometer-resolution serial section EM. The dataset spans the brain and anterior spinal cord, capturing >180,000 segmented soma, >40,000 molecularly annotated neurons, and 30 million synapses, most of which were classified as excitatory, inhibitory, or modulatory. To characterize the directional flow of activity across the brain, we leverage the synaptic and cell body annotations to compute region-wise input and output drive indices at single cell resolution. We illustrate the dataset’s utility by dissecting and validating circuits in three distinct systems: water flow direction encoding in the lateral line, recurrent excitation and contralateral inhibition in a hindbrain motion integrator, and functionally relevant targeted long-range projections from a tegmental excitatory nucleus, demonstrating that this resource enables rigorous hypothesis testing as well as exploratory-driven circuit analysis. The dataset is integrated into an open-access platform optimized to facilitate community reconstruction and discovery efforts throughout the larval zebrafish brain.
Acknowledgements
F. Engert received funding from the National Institutes of Health (U19NS104653 and 1R01NS124017-01), DoD (W911NF2420112) and the Simons Foundation (SCGB 542973 and NC-GB-CULM-00003241-02. J. W. Lichtman received funding from the National Institutes of Health (U24NS109102, U19NS104653, and UM1NS132250). This project was partially supported by NIH grants 1U01NS132158 and R01HD104969 to H. Pfister, Simons Foundation SCGB 542943SPI to M. Ahrens and by the Howard Hughes Medical Institute. We are grateful to F. Collman, D. Brittain, A. Halageri, and S. Dorkenwald for constructive feedback and advice on the CAVE deployment. P. Petkova contributed critical segmentation of both LM and EM datasets and generated manual segmentation ground truth, for which we are especially grateful. We thank V. Wang and A. Chen for providing example neurons used to test the polarity classifier. We also acknowledge B.C. Oban and T. Skye for maintaining a supportive working environment. Finally, we thank J. Miller and K. Hurley for their expert care and management of the fish facility, and E. Nako and S. Montgomery for their administrative support.